Table Of Content

Bisulfite PCR is a multi-step process that enables users to determine the methylation status of CpG sites within a PCR amplicon. The first step in bisulfite PCR is bisulfite conversion of the DNA. Bisulfite conversion is a process that uses sodium bisulfite to convert all unmethylated cytosines in DNA to uracil. In contrast, methylated cytosines are left unaltered by the sodium bisulfite treatment. After the bisulfite conversion step, a PCR reaction is run to determine the methylation status of CpG sites located between the two primers. During this PCR, the converted uracil bases are read and amplified as thymine, while the methylated cytosines continue to be amplified as cytosine.
Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies
The maximum number of candidate primer pairs to screen in order to find specific primer pairs (The candidate primers are generated by primer3 program). Increasing this number can increase the chance of finding a specific primer pair but the process will take longer. In order to obtain RT-qPCR results with high accuracy, each step must be performed satisfactorily in the preanalytical, analytical, and post-analytical steps. It is important to avoid sample contamination and ensure adequate procedures for specimen collection, handling, transport, and storage.
Steps
While there was more variability in the amount of product formed at increasing concentrations of MgCl2, the most PCR product was observed at 4 mM Mg2+ (lane 9), the same concentration observed for the yeast GAL3 gene. The program will use the RefSeq mRNA sequence from the organism you selected to design the primers. To avoid amplification of contaminating genomic DNA, design primers so that one-half of the primer hybridizes to the 3′ end of one exon, and the other half to the 5′ end of the adjacent exon. The NCBI tool Primer-BLAST is widely used for qPCR primer design.
References
This can take a while but the screen updates periodically to show the time since submission. Avoid complementarity that can allow two primers to hybridize at their 3´ ends. The unintended outcome is that a polymerase can then extend to form dimerized PCR products. Much the same, avoid tangling up your reaction primers with hairpin loop secondary structures within individual primers. DNA polymerases can be slowed down by such thermo-stable secondary structures. While it’s usually not needed, keep in mind that primer degradation by an enzyme’s proofreading activity will be inhibited by incorporating phosphorothioate linkages into the 2 bases at the oligos 3´ end.
Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using ... - Nature.com
Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID- using ....
Posted: Tue, 16 Jun 2020 07:00:00 GMT [source]
The millimolar concentration of deoxyribonucleotide triphosphate. This argument is considered only if Concentration of divalent cations is specified.
A tool to automatically design multiplex PCR primer pairs for specific targets using diverse templates
Even if the same tools are used, the same suggested primers and probes may not always be the same, but this does not indicate that the quality of primer design is poor. For example, the primers and probes designed for the ORF 1ab of SARS-CoV-2 have different sequences distributed by the Chinese CDC and the USA CDC. Indeed, there are many primer and probe combinations that can meet the requirements of RT-qPCR experiments, so the highest quality combination does not always need to be used. The amplification conditions also need to be continuously optimized through experiments. For example, the optimal Tm and optimized oligo concentrations were eventually confirmed by temperature gradient experiments. The parameters given by the design tool are for reference only and should never replace the subsequent confirmation process.
Plate Format Ordering at IDT DNA Oligos
Genes from the budding yeast Saccharomyces cerevisiae and from an uncharacterized Mycobacteriophage were amplified in these experiments. The standard 3-step PCR protocol outlined in Table 2 was employed for all three experiments described below. This section covers some of the basic settings for your primers, including the PCR product size and melting temperature. There is also an option to include sequences for reverse or forward primers. The first step in designing primers is to get the nucleotide sequence of your gene of interest.
PrimerSuite: A High-Throughput Web-Based Primer Design Program for Multiplex Bisulfite PCR Scientific Reports - Nature.com
PrimerSuite: A High-Throughput Web-Based Primer Design Program for Multiplex Bisulfite PCR Scientific Reports.
Posted: Tue, 24 Jan 2017 08:00:00 GMT [source]
IVT Product Purification
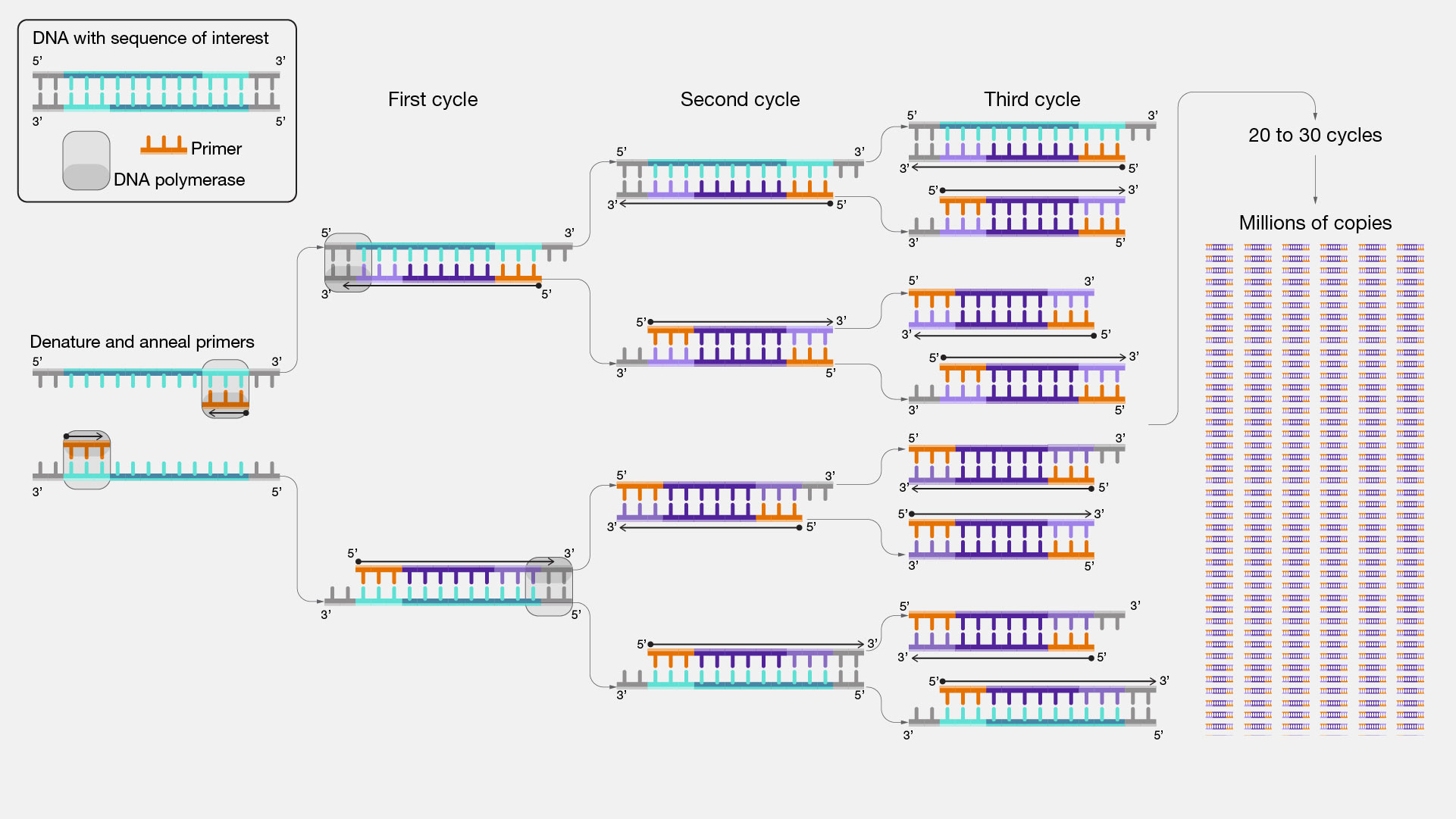
In a PCR experiment, usually two primers are used in each experiment. Each of the two primers (forward primers and reverse primers) are designed in such a way that they flank the target region of the gene sequence to be copied on the template DNA molecule (Figure 1). Notice that in the experiments presented in Figures 3A and 3B, a discrete band was obtained using the cycling conditions thought to be optimal based on primer annealing temperatures.
From an economic point of view, it is generally recommended to design a probe and three corresponding pairs of primers for a target and determine the best primer combination after experimental screening. The entire flow of the primer design is shown in Figure Figure44. Primer-BLAST offers a number of features that are not available in other software tools. Table 1 gives a brief summary of these features, many of which are important for various primer design requirements and allows examination of primer specificity details by users.
Cross-check if the sequence has been reverse complemented or not. Although the names suggest they create copies of different strands, their names depend on the direction of the strand being used for amplification. Conceived the idea for STITCHER 2.0 and wrote the code and designed the website shown in Figure 1. This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Before reverse complementation the sequence is 5’-CTGGAGGACGGAAGAGGAAGTAA-3’ and after reverse complementation 5’-TTACTTCCTCTTCCGTCCTCCAG-3’. Image showing the steps involved in reverse complementation in APE software. We are using APE (A Plasmid Editor) software to design the primers, which is free to use. When Primerize gives warnings about misprimings, we carry out both the one-shot protocol above and the following multiple-round strategies as well, evaluating attainment of full-length DNA template by agarose gel. This a sequence that we repeatedly re-use as a primer binding site, which we call ‘Tail2’. It is critical (see below) to check in silico that this tail has negligible likelihood of base pairing with your sequence of interest.


No comments:
Post a Comment